Types
The MASS Toolbox relies heavily on data types and custom notation to facilitate the process of model construction, manipulation, and analysis.
| Out[2]= | 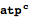 |
| Out[2]= | 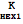 |
It is very important to make clear that although metabolites (and many of the following data types) are displayed in a human readable format (e.g
atpc), the internal representation is conserved,
Out[3]//FullForm= |
| | 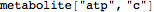 |
and is in this respect very similar to other
Mathematica definitions, e.g.,
Out[4]//FullForm= |
| | 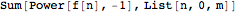 |
It is possible to use "Evaluation-> Evaluate in place" in the menu (or Cmd + Shift + Enter or the mouse context menu) for inplace replacements (select the next line and try it)
Select the following code and hit Cmd + Shift + Enter (metabolite["atp","cytosol"] should be converted to
atpcytosol)
| Out[6]= | 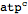 |
| Out[8]= | 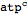 |
Species
In order to define a model of biochemical process one has to define the participating species. The Toolbox provides a series of species type definitions.
| species["id"] | a species |
| species["id", "compartment"] | a species in a particular compartment |
| species["id", _] | a species in any compartment |
| metabolite["id", "compartment"] | a small compound in a particular compartment |
| m["id", "compartment"] | shorthand for metabolite["id", "compartment"] |
| enzyme["ID"→"id", "Compartment"→"compartment",opts] | a enzyme in a particular compartment |
Toolbox species types
This defines adenosine triphosphate (ATP) in the cytosol
| Out[35]= | 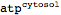 |
This defines adenosine triphosphate (ATP) using a slightly shorter notation
| Out[36]= | 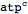 |
However, the full type expression (metabolite) is used internally
Out[37]//FullForm= |
| | 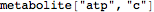 |
Reactions
Chemical reactions are defined in the following way:
| reaction["id", substrates, products, stoichiometry] | a reversible reaction |
| reaction["id", substrates, products, stoichiometry, False] | an irreversible reaction |
| r[args] | a shorthand for reaction |
Toolbox reactions
| Out[38]= | 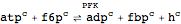 |
Define an irreversible reaction
| Out[38]= | 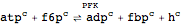 |
Query data types
Toolbox typical accessor function
Query the ID and compartment of a metabolite
| Out[45]= | 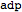 |
| Out[46]= | 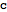 |
Query the ID and compartment of an reaction
| Out[42]= | 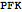 |
| Out[43]= | 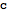 |
Textual input
| str2mass["ID"] | a metabolite (ID) |
| str2mass["ID[Comp]"] | a metabolite in a particular compartment (IDComp) |
| str2mass["ID: 2 x <=> y + z"] | a reversible reaction (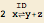 ) ) |
| str2mass["ID: 2 x[c] --> y[c] + z[c]"] | an irreversible reaction (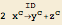 ) ) |
| str2mass["Keq_ID"] | an equilibrium constant for reaction ID (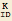 ) ) |
| str2mass["k_ID_fwd"] | forward rate constant |
| str2mass["k_ID_rev"] | reverse rate constant |
Use textual input to declare species, parameters, reactions etc.
| Out[54]= | 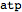 |
Out[55]//FullForm= |
| | 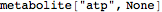 |
Parse a metabolite with a specific compartment
| Out[56]= | 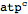 |
Out[57]//FullForm= |
| | 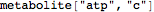 |
Facilitate
str2mass to quickly define a model
| Out[90]= | 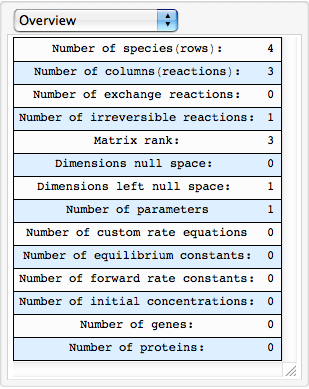 |
| Out[91]= | 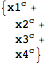 |